Documents
Poster
Compressive Regularized Discriminant Analysis of High-Dimensional Data with Applications to Microarray Studies
- Citation Author(s):
- Submitted by:
- Muhammad Naveed...
- Last updated:
- 13 April 2018 - 12:03am
- Document Type:
- Poster
- Document Year:
- 2018
- Event:
- Presenters:
- Muhammad Naveed Tabassum
- Paper Code:
- SPTM-P2.1
- Categories:
- Keywords:
- Log in to post comments
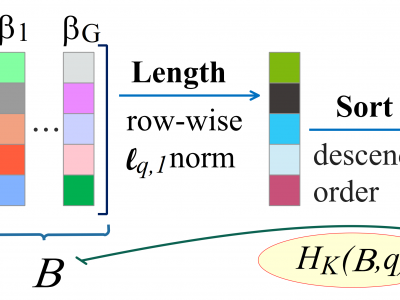
We propose a modification of linear discriminant analysis, referred to as compressive regularized discriminant analysis (CRDA), for analysis of high-dimensional datasets. CRDA is specially designed for feature elimination purpose and can be used as gene selection method in microarray studies. CRDA lends ideas from ℓq,1 norm minimization algorithms in the multiple measurement vectors (MMV) model and utilizes joint-sparsity promoting hard thresholding for feature elimination. A regularization of the sample covariance matrix is also needed as we consider the challenging scenario where the number of features (variables) is comparable or exceeding the sample size of the training dataset. A simulation study and four examples of real-life microarray datasets evaluate the performances of CRDA based classifiers. Overall, the proposed method gives fewer misclassification errors than its competitors, while at the same time achieving accurate feature selection.

