Documents
Presentation Slides
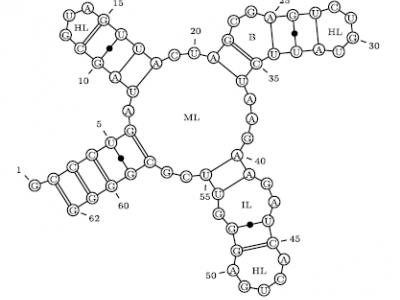
RNA secondary structures: from ab initio prediction to better compression, and back
- Citation Author(s):
- Submitted by:
- Evarista Onokpasa
- Last updated:
- 3 March 2023 - 5:52am
- Document Type:
- Presentation Slides
- Document Year:
- 2023
- Event:
- Presenters:
- Evarista Onokpasa
- Paper Code:
- 235
- Categories:
- Keywords:
- Log in to post comments
In this paper, we use the biological domain knowledge incorporated into stochastic models
for ab initio RNA secondary-structure prediction to improve the state of the art in joint
compression of RNA sequence and structure data (Liu et al., BMC Bioinformatics, 2008).
Moreover, we show that, conversely, compression ratio can serve as a cheap and robust
proxy for comparing the prediction quality of different stochastic models, which may help
guide the search for better RNA structure prediction models.
Our results build on expert stochastic context-free grammar models of RNA secondary
structures (Dowell & Eddy, BMC Bioinformatics, 2004; Nebel & Scheid, Theory in Biosciences, 2011)
combined with different (static and adaptive) models for rule probabilities
and arithmetic coding. We provide a prototype implementation and an extensive empirical evaluation,
where we illustrate how grammar features and probability models affect
compression ratios

